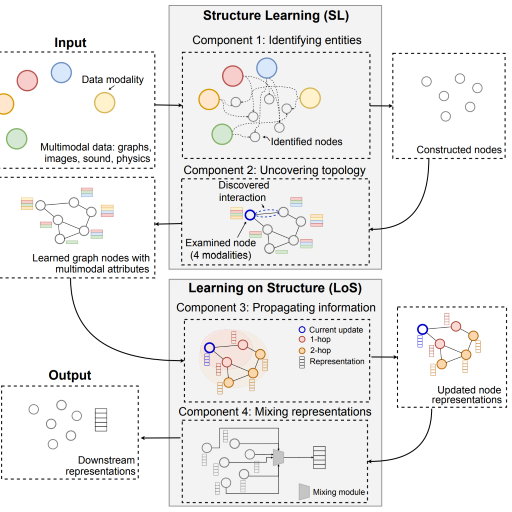
Multimodal Learning with Graphs
Survey of 140 studies in graph-centric AI to establish a framework for multimodal graph learning.

Hi! 👋🏽 I am a DPhil (AKA, PhD) student at the University of Oxford and a Rhodes Scholar. Motivated by personal experiences, I conduct research at the interface of artificial intelligence (AI), translational neuroscience, and precision medicine. I seek to develop AI technologies that advance the frontier of personalized diagnosis and treatment for neurological disorders and other challenging medical conditions.
📖 Education. I have over eight years of research training, advised by:
I graduated from Harvard University with a joint Bachelor’s in Computer Science and Neuroscience as well as a concurrent Master’s in Computer Science. There, I was named a John Harvard Scholar, received the Paul Revere Frothingham Scholarship Prize, and was elected to Harvard’s chapter of Phi Beta Kappa. I previously attended Phillips Exeter Academy.
💡 Research. My research efforts have produced over 30 papers – including seven first or co-first author works – in peer-reviewed journals including Cell, Nature Neuroscience, Nature Machine Intelligence, Nature Aging, npj Digital Medicine, Alzheimer's & Dementia, JMIR, Journal of Neuroinflammation, and Neurobiology of Disease. My work has been featured in 17 posters at international conferences (e.g., SfN, GEM and SLLM at ICLR, AAIC, Glia, AAN, IDWeek, ML4H at NeurIPS, AD/PD, AAIC Neuroscience Next) and I have presented 24 posters or talks at local symposia, receiving the Neurology Department Award at MGH Clinical Research Day.
🧬 Fellowships and grants. I have received 21 fellowships or grants, including the Encode: AI for Science PhD Fellowship, Y Combinator Summer Fellows Grant, Barry Goldwater Scholarship, Undergraduate Technology Innovation Fellowship at Harvard Business School, Yun Family Research Fellowship for Revolutionary Thinking, Herchel Smith Undergraduate Science Research Fellowship, TIME Initiative Fellowship, and Coca-Cola Scholarship.
🍎 Leadership and teaching. I previously co-founded the Harvard Undergraduate OpenBio Laboratory; served as Operations Director of the BIDMC Crimson Care Collaborative Clinic, a student-faculty teaching clinic providing outpatient primary care at Beth Israel Deaconess Medical Center; and was Chair of the Demos and Perspectives Committee at the 2024 Machine Learning for Health Conference. I am devoted to teaching and mentorship: at Harvard, I was Co-Captain of Peer Concentration Advising in Computer Science, Peer Advising Fellow, and Pre-Health Peer Advisor.
I am excited about translating our academic discoveries from bench to bedside. When not coding, reading, or brainstorming with founders, I love creative writing. My writing on neurological disease was named as the New York Times Gold Medal Writing Portfolio by the Scholastic Art & Writing Awards, and I was a finalist in the 89th Scripps National Spelling Bee. I also enjoy photography, bird watching, and meditation.
Some of my work is shown below. If anything piques your interest, I would love to hear from you. Please don’t hesitate to reach out!
First and co-first author publications are indicated in red. For a complete and current list of my publications, please see Google Scholar.

Survey of 140 studies in graph-centric AI to establish a framework for multimodal graph learning.
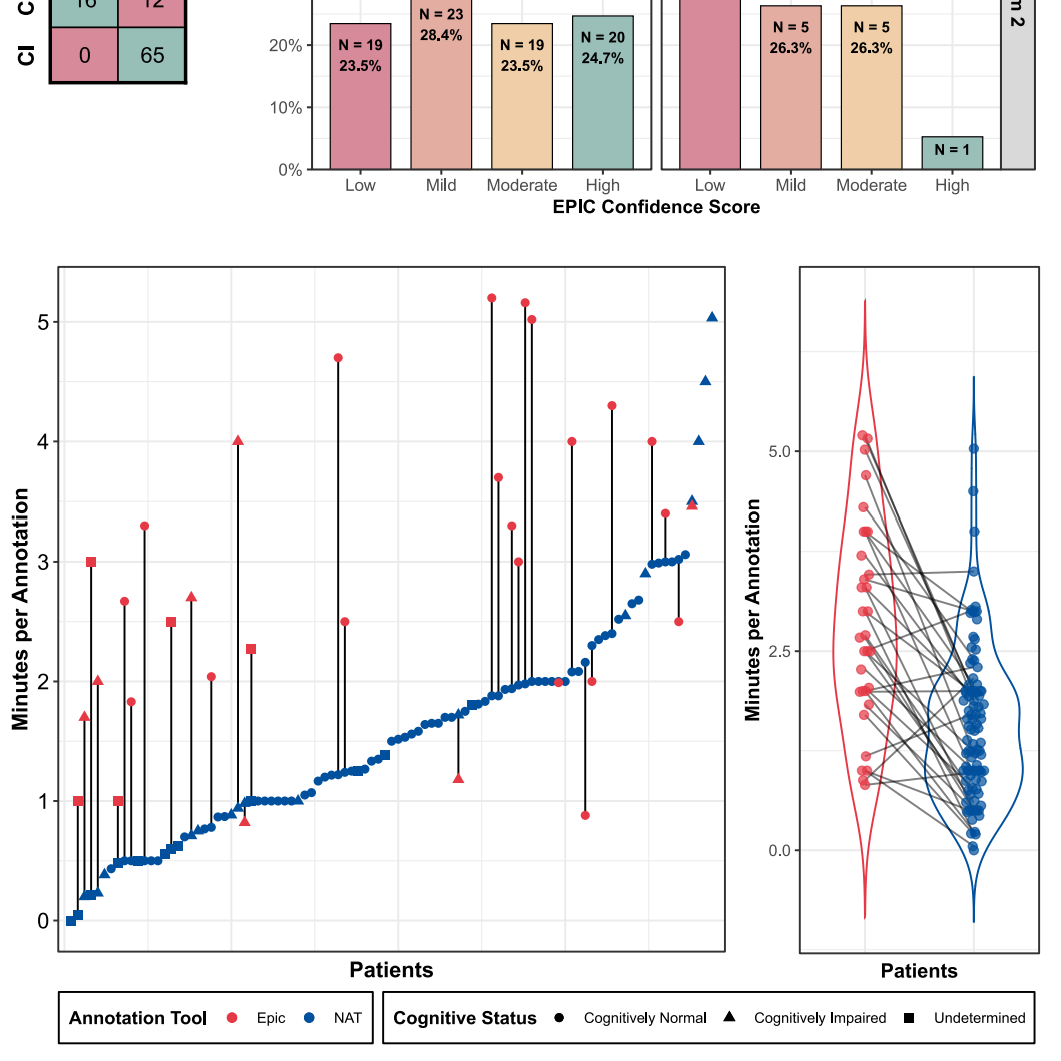
Development and evaluation of a natural language processing annotation tool to facilitate phenotyping of cognitive status in electronic health records.

Plasma biomarkers for prognosis of cognitive decline in patients with mild cognitive impairment.
Cyclic multiplex fluorescent immunohistochemistry and machine learning reveal distinct states of astrocytes and microglia in normal aging and Alzheimer’s disease.
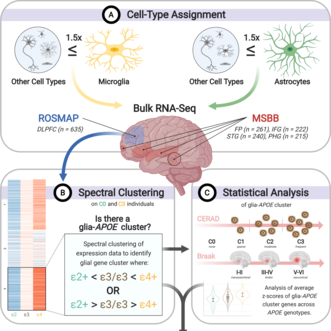
Effect of APOE alleles on the glial transcriptome in normal aging and Alzheimer’s disease.
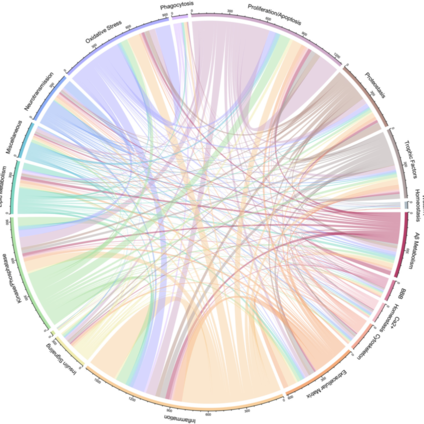
Systematic review of human post-mortem immunohistochemical studies and bioinformatics analyses unveil the complexity of astrocyte reaction in Alzheimer's disease.
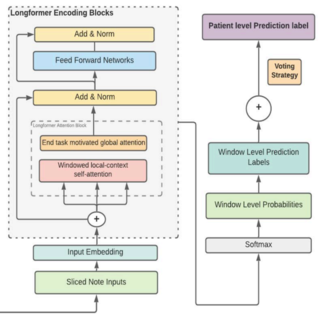
Active deep learning to detect cognitive concerns in electronic health records.
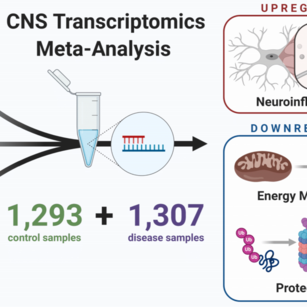
Systematic review and meta-analysis of human transcriptomics reveals neuroinflammation, deficient energy metabolism, and proteostasis failure across neurodegeneration.
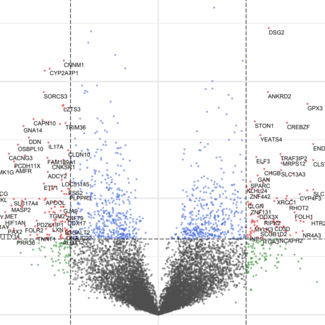
Differential gene expression data from the human central nervous system across Alzheimer's disease, Lewy body diseases, and the amyotrophic lateral sclerosis and frontotemporal dementia spectrum.
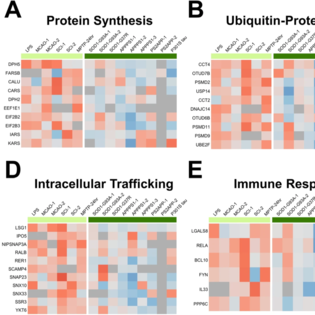
Meta-analysis of mouse transcriptomic studies supports a context-dependent astrocyte reaction in acute CNS injury versus neurodegeneration.
Composition of Caenorhabditis elegans extracellular vesicles suggests roles in metabolism, immunity, and aging.
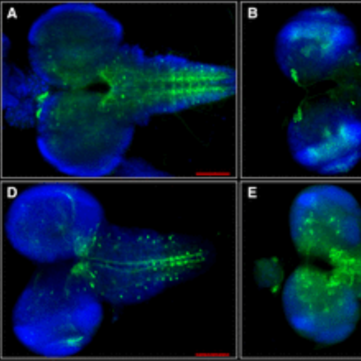
An interscholastic network to generate LexA enhancer trap lines in Drosophila.

Advised by Dr. Marinka Zitnik in the Department of Biomedical Informatics at Harvard Medical School, studying applied machine learning on network systems in biology and medicine.

Advised by Dr. George Church and Dr. Jenny Tam, applying graph AI on patient-derived cerebral organoids for drug repurposing in neuropsychiatric disorders.

Advised by Dr. Sudeshna Das at the MassGeneral Institute for Neurodegenerative Disease, developing integrative computational methods in biomedical and brain research.

Was previously Biology Fellow at Fifty Years, helping great scientists leverage breakthroughs in biology to found iconic companies and solve pressing global challenges.

Led partnerships at Nucleate Dojo to empower the next-generation of biotechnology innovators.
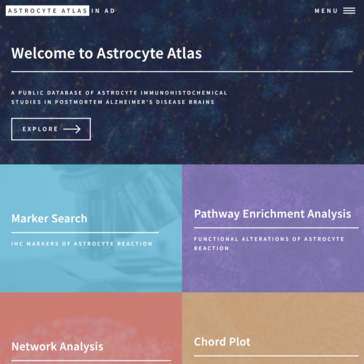
A publicly-available atlas of markers of reactive astrogliosis in Alzheimer's and associated bioinformatics analyses.

Open data analysis platform for Alzheimer's disease.
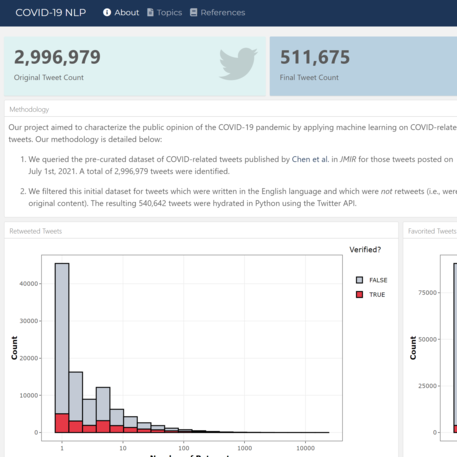
For the Harvard Data Analytics Group Data Science Fellowship, characterized the public pandemic discourse by applying NLP on COVID-related tweets.